Usage
4.1 Basic usage
1. Format of input data
SoCube's data input is a scRNA-seq UMI count matrix with rows of cells, columns of genes, and values of UMI counts. SoCube accepts two data formats: (1)DataFrame objects in HDF5 format saved by Python pandas library; (2)AnnData objects in H5AD format (a customized HDF5 format) saved by Python anndata library. People working in the field of single-cell omics have some basic knowledge of data processing, so this manual does not specifically describe how to convert from other formats to the above formats.
2. Running SoCube
SoCube runs under the terminal like the aforementioned pip. The advantage is that it can run on many servers without GUI. Assuming that the input file is "D:\data\pbmc-1C-dm.h5ad", you can enter the following command in the terminal to run the software, and then you will see the software start and output logs in the terminal. The parameter -i (or --input) indicates the specified input file.
socube --input "D:\data\pbmc-1C-dm.h5ad"
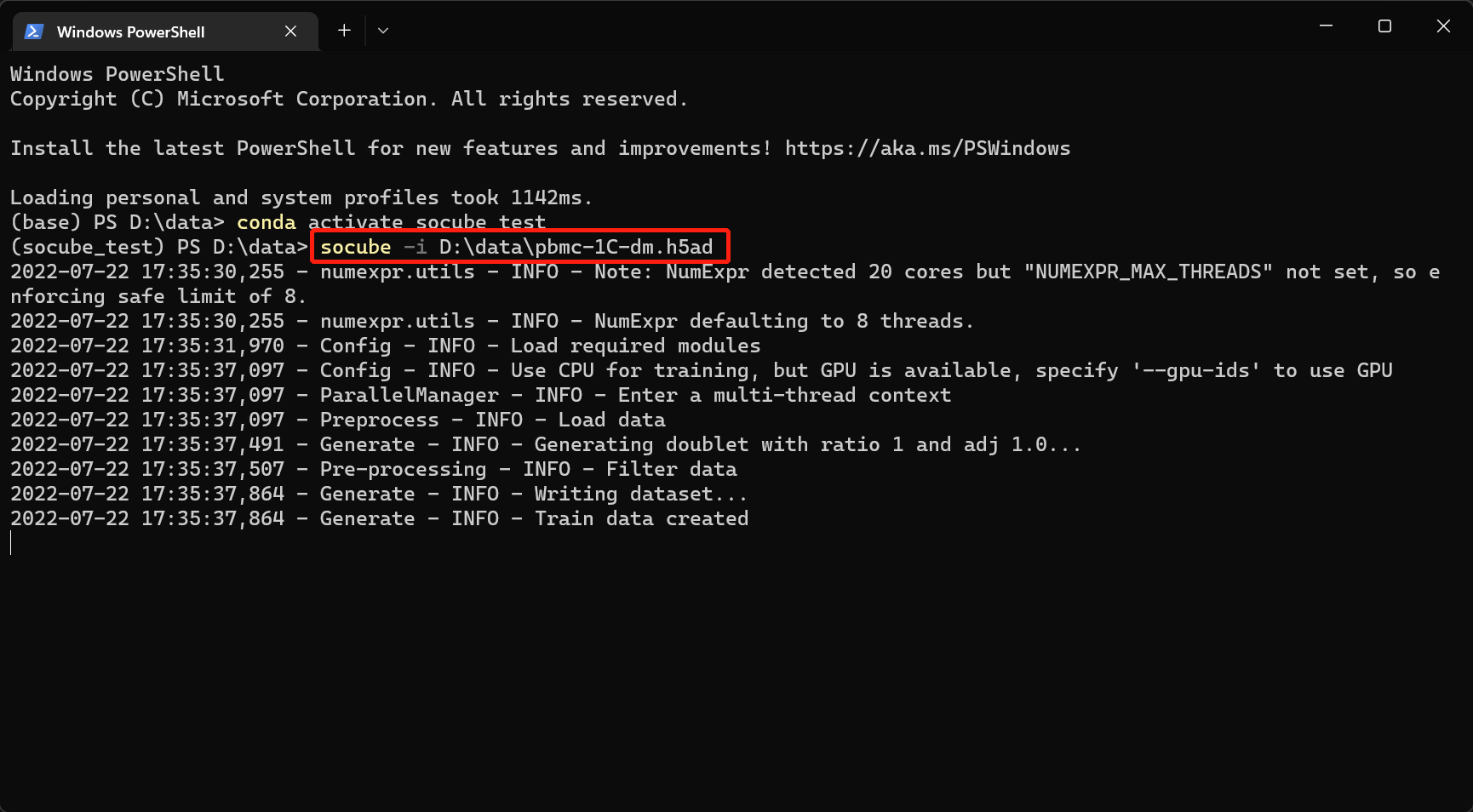
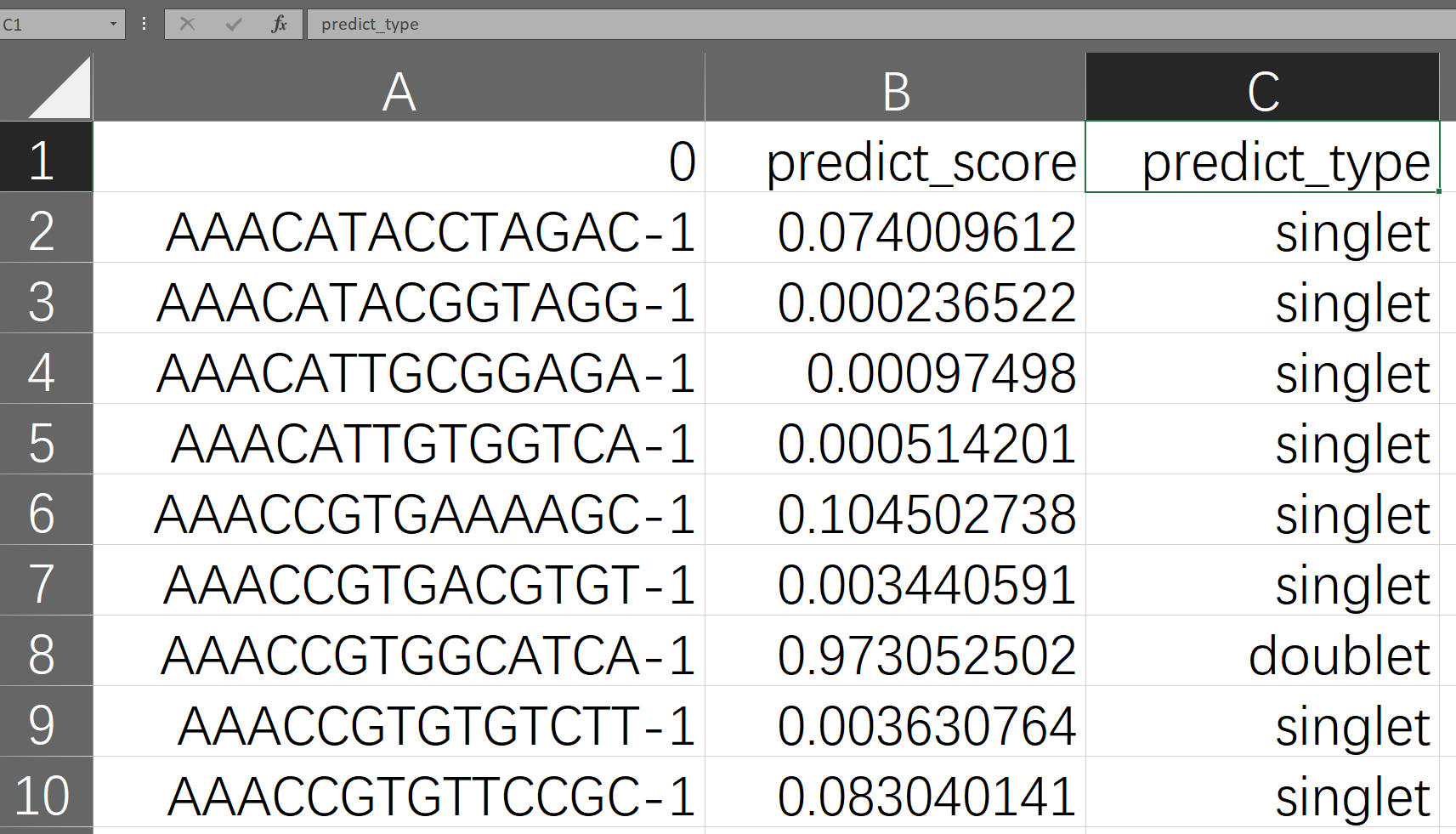
When the process is finished, the doublet detection results and the intermediate files will be put together with the input files by default, folders named as embedding, plots, models, and outputs are created. The final results are in a CSV file starting with "final_result" in the "outputs/your model ID/" folder. Open it with Excel spreadsheet software, and you can see that it has three columns as shown below. The first column is the name of the cell sample, determined by the user's input; the second column is the probability value of SoCube predicting the sample to be a doublet, ranging from 0 to 1; the third column is SoCube's prediction of the type based on the probability threshold given by the user or the default threshold of 0.5. Users can either use the probability values for subsequent custom filtering or use the third column for direct filtering.
4.2 GPU accelerated computing
If you has a discrete NVIDIA graphics card and the correct CUDA configuration, you will receive the message "Using CPU training, but GPU is available, specify '-gpu-ids ' to use GPU". You only need to add the parameter "-gpu-ids" to the previous command to specify the GPU to be used. The GPU ID can be seen in the output of the nvidia-smi command mentioned earlier, 0,1,2,... which means the 1st, 2nd, 3rd... GPUs respectively (in the case of multiple GPUs)
socube -i "D:\data\pbmc-1C-dm.h5ad" --gpu-ids "0,1"
4.3 Docker-based usage
The docker-based usage is similar to the previously mentioned usage after pip installation, but requires docker to start the container, so there are additional steps docker run to start the container and docker exec to enter the container. "-v", "--gpus", "--name" are all start parameters of docker run command, " -v" parameter mounts the external folder to the internal path of the docker container, because socube reads the file path inside the docker container. The "--gpus" parameter is responsible for authorizing the number of gpu used by the container.
# create container
docker run -v D:/data:/workspace/datasets `
–gpus all `
–name socube `
gcszhn/socube:latest
# enter container
docker exec -it soube bash# create container
sudo docker run -v /data:/workspace/datasets \
–gpus all \
–name socube \
gcszhn/socube:latest
# enter container
sudo docker exec -it soube bash"gcszhn/socube:latest" is the image name. The parameters before it are docker startup parameters, including folder mapping and GPU mount, and the parameters after it are SoCube parameters, which are used in the same way as before. Users can check the image name through docker images.
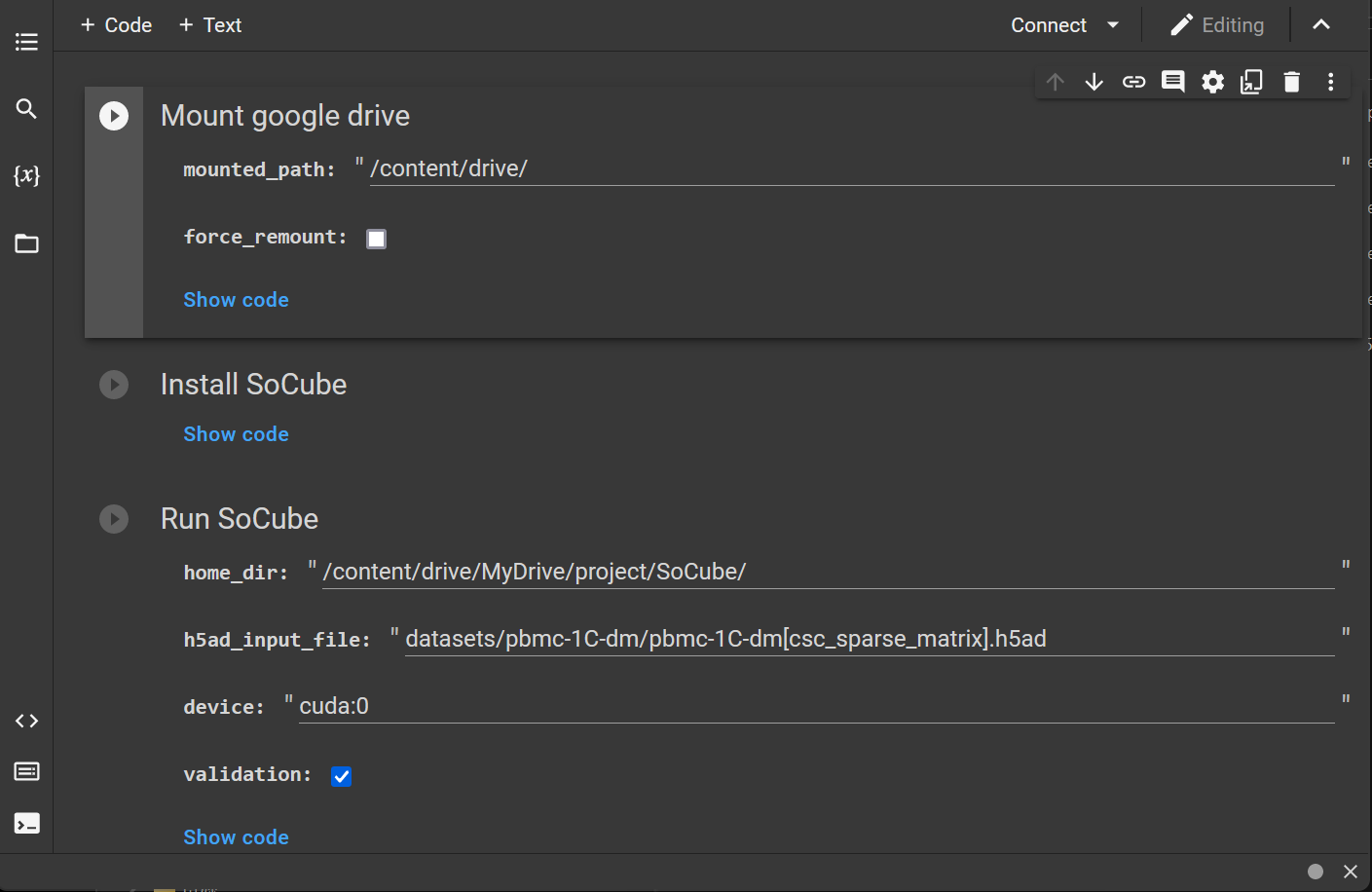
4.4 Usage of colab
Google offers a free online machine learning platform with GPU colab. Users can upload socube_colab.ipynb (available in the open source repository of this project) and scRNA-seq data to your google drive and use it for GPU-accelerated prediction. (Tip: you need to choose to enable GPU inside the notebook settings, the default is CPU).
4.5 Multi-process training
With sufficient memory and video memory, users can use the --enable-multiprocess parameter to enable multi-process training acceleration to take full advantage of the multi-core computing of modern CPUs.
socube -i your_sc.h5ad -o your_sc --gpu-ids 0 --enable-multiprocess